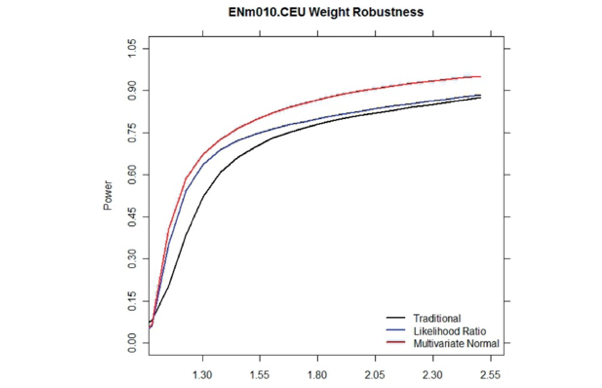
ML for Health
Machine learning scientist specializing in computational biology and healthcare AI
I am currently a Staff Machine Learning Scientist on the clinical machine learning team at insitro, where I develop multi-modal genomic and clinical models to discover and validate novel genetic mechanisms of disease and to prioritize therapeutic targets.
Prior to insitro, I was a Senior Research Scientist on the Health AI team at Apple, developing machine learning methods deployed across Apple's health ecosystem to derive insights into human physiology from sensor signal data.
I completed my postdoctoral fellowship at the Institute for Computational and Experimental Research in Mathematics (ICERM) at Brown University, participating in the semester program on model and dimension reduction in uncertain and dynamic systems. During this fellowship, I collaborated closely with Sohini Ramachandran, Lorin Crawford, Sigal Gottlieb, and Yanlai Chen.
I earned my PhD in machine learning and statistical genetics through Princeton University's Quantitative and Computational Biology program, where I was advised by Barbara Engelhardt (Princeton) and co-advised by Sayan Mukherjee (Duke). During my graduate studies, I gained experience in industry and academic research labs, including Microsoft Research New England (with Jennifer Listgarten and Nicolo Fusi), Rockefeller University (with Robert Darnell and Chaolin Zhang), and Harvard School of Public Health (with Alkes Price). My undergraduate research mentor at UCLA was Eleazar Eskin.
Education
- Princeton University
Ph.D. Quantitative and Computational Biology (2014-2019)
- UCLA
B.S. Computer Science, minor Bioinformatics (2008-2013)
Research

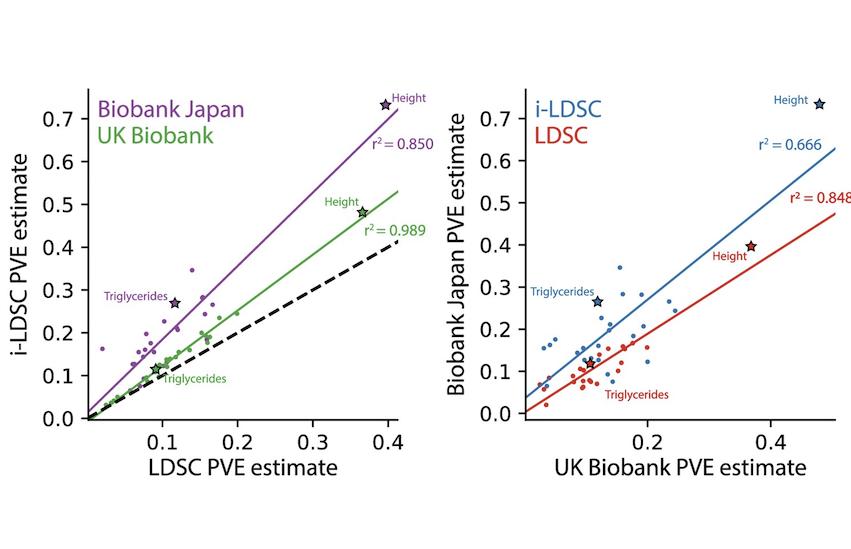
Discovering non-additive heritability using additive GWAS summary statistics
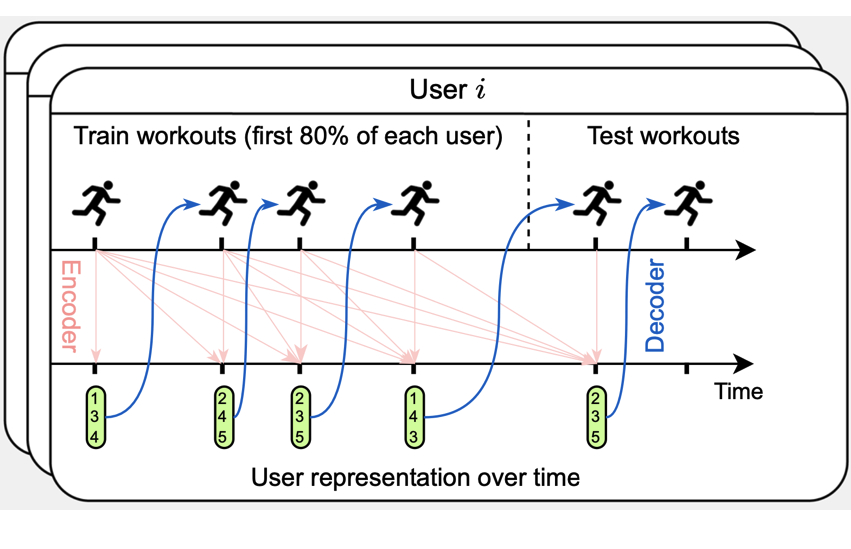
Modeling personalized heart rate response to exercise and environmental factors with wearables data
Latent Temporal Flows for Multivariate Analysis of Wearables Data
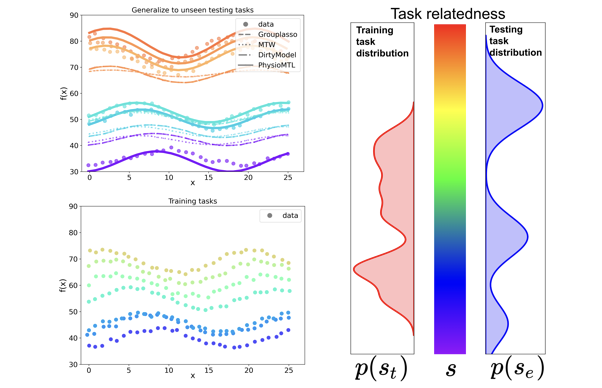
PhysioMTL: Personalizing Physiological Patterns using Optimal Transport Multi-Task Regression
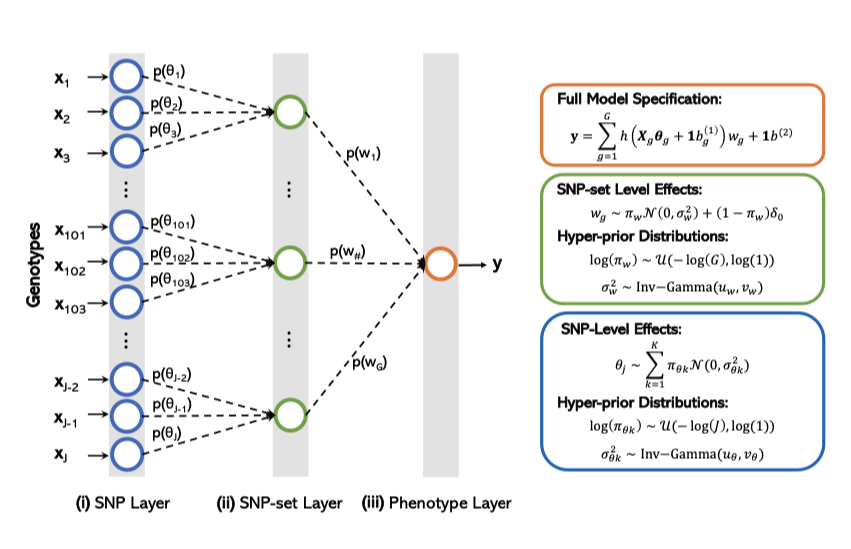
Multi-scale Inference of Genetic Trait Architecture using Biologically Annotated Neural Networks
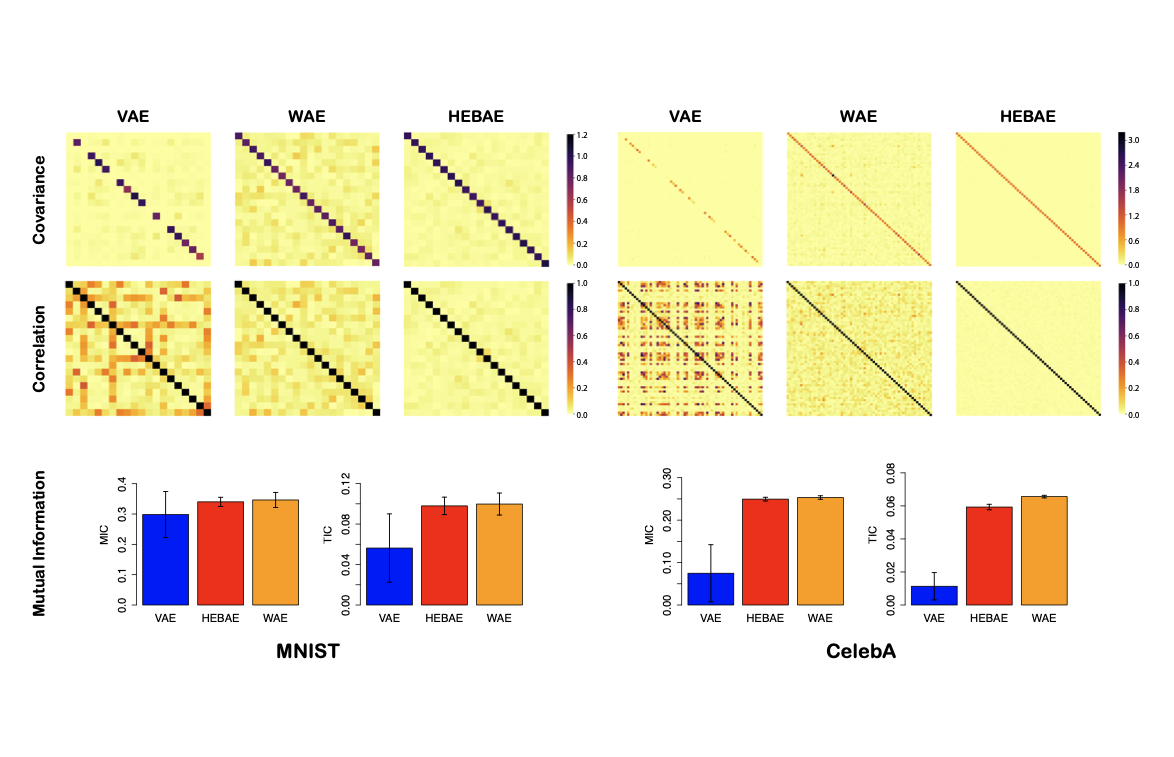
Generalizing Variational Autoencoders with Hierarchical Empirical Bayes
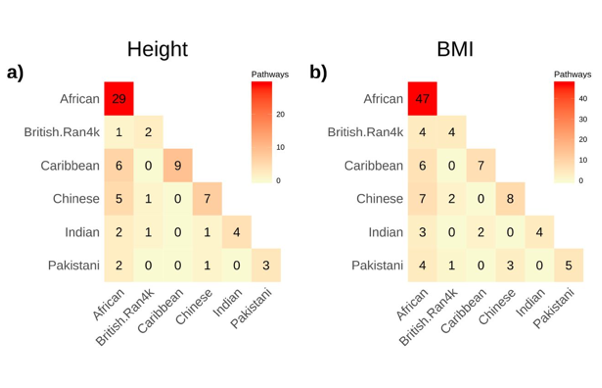
Pathway Analysis within Multiple Human Ancestries Reveals Novel Signals for Epistasis in Complex Traits
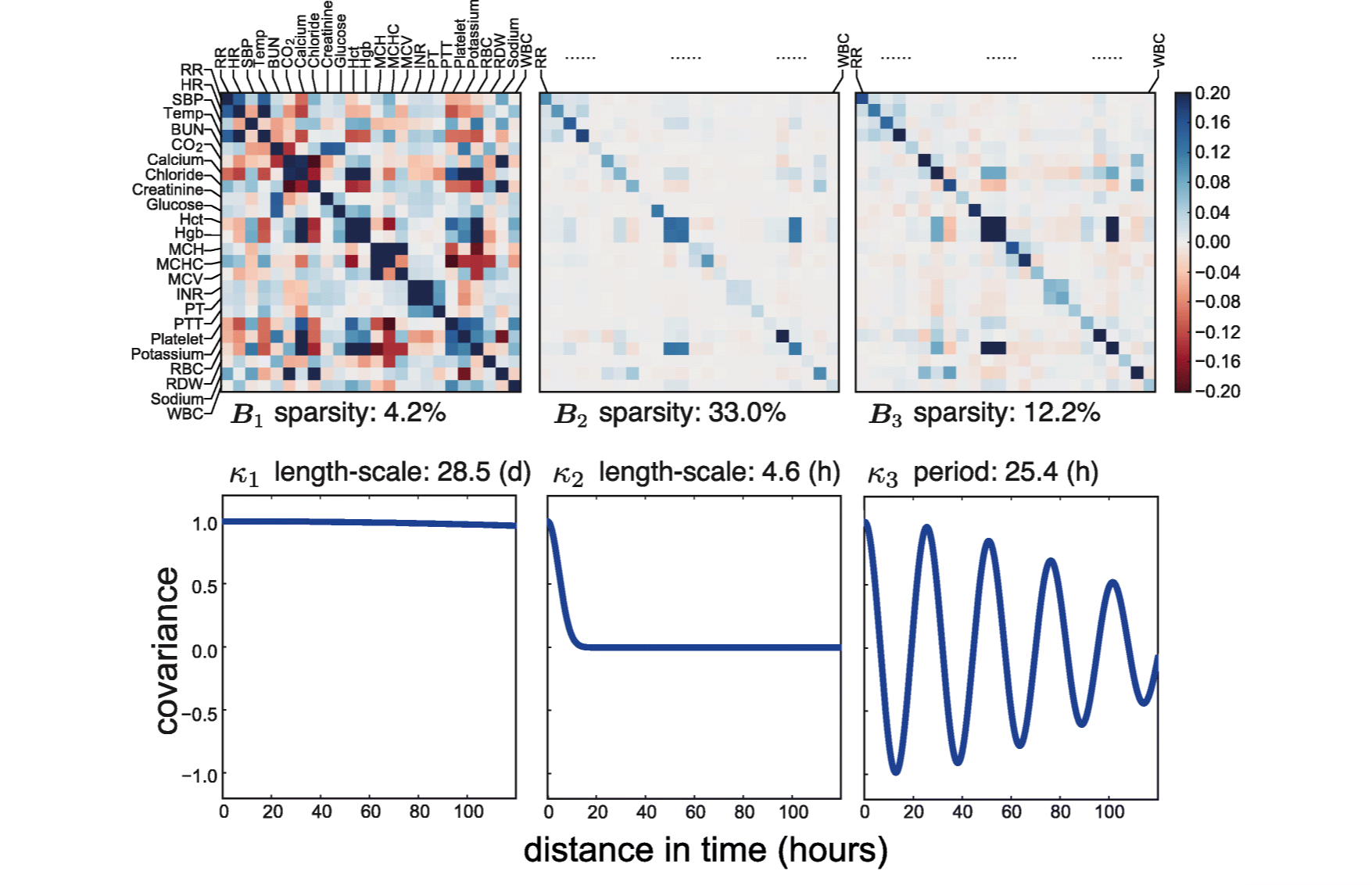
Sparse multi-output Gaussian processes for online medical time series prediction
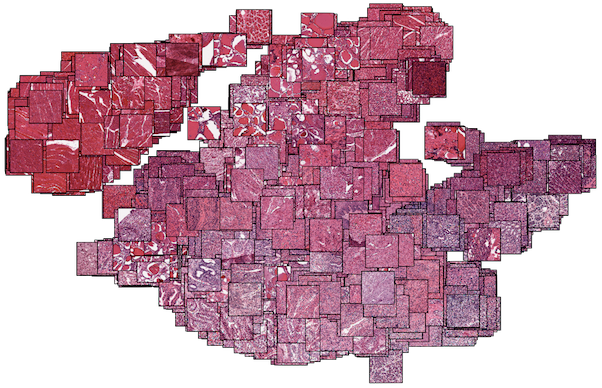
Joint analysis of gene expression levels and histological images identifies genes associated with tissue morphology
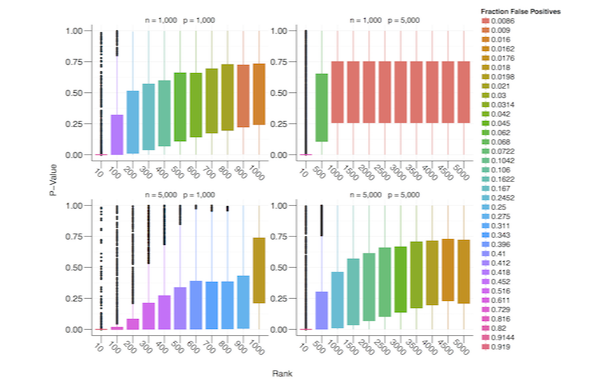
Adaptive Randomized Dimension Reduction on Massive Data
Statistical tests for detecting variance effects in quantitative trait studies